Research
The Cheeseman lab seeks to define the molecular mechanisms that govern cellular processes, with a particular focus on cell division and its regulation. Our research combines cutting-edge microscopy, biochemistry, and genomics approaches to understand how cells coordinate complex events throughout the cell cycle. By integrating high-throughput screening methods with focused mechanistic studies, we aim to understand both normal cellular function and its disruption in disease states.
Biological Discovery Through Large-Scale Functional Genomics Screens
We develop and implement large-scale screening approaches that combine CRISPR-based gene targeting with sequencing and automated microscopy. These large-scale analyses allow us to systematically uncover the functions of understudied genes and place known genes in new biological contexts. Our targeted screening approaches focus on specific biological questions, enabling deep insights into cellular processes including cell division, quiescence, and drug sensitivity. Through this work, we aim to build comprehensive resources for the cell biology community while advancing our understanding of fundamental cellular mechanisms.
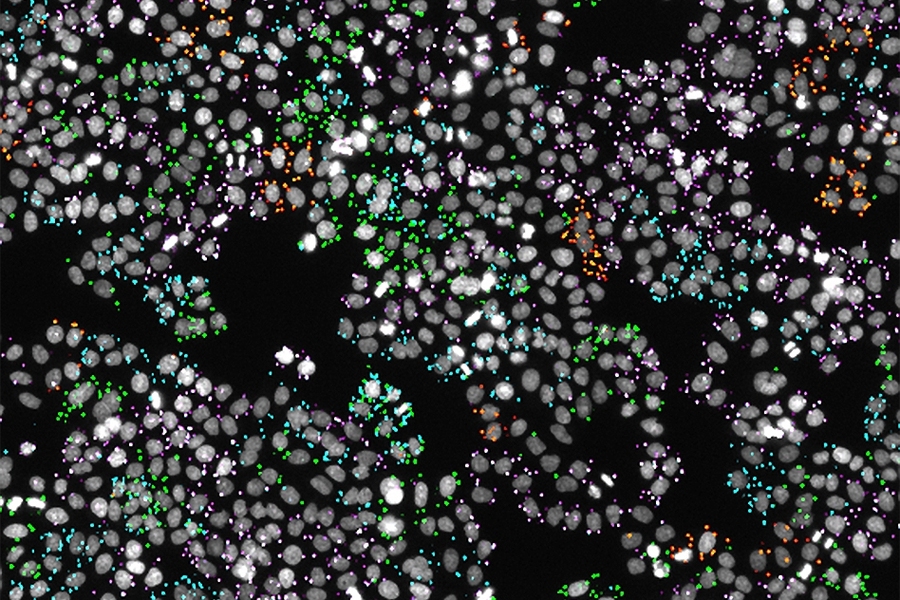
Recent Publications
Di Bernardo M, Kern RS, Mallar A, Nutter-Upham A, Blainey PC, Cheeseman IM
bioRxiv (2025)
Su KC, Radul E, Maier NK, Tsang MJ, Goul C, Moodie B, Marescal O, Keys HR, Cheeseman IM
Journal of Cell Biology (2025)
Funk L, Su KC, Ly J, Feldman D, Singh A, Moodie B, Blainey PC, Cheeseman IM
Cell (2022)
Control of Gene Expression in Different Cell States
We investigate how cells regulate their gene expression programs across the cell cycle, in different cellular states, and in response to various stresses. Using a combination of transcriptomics, proteomics, and advanced imaging techniques, we aim to understand the regulatory mechanisms that govern transitions between cell states. Our research extends from fundamental cell cycle regulation to specialized states, such as meiosis, quiescence, and senescence, with implications for development and disease. We are particularly interested in understanding how different layers of regulation, from transcription and splicing to translational control and post-translational modification, coordinate to define distinct cellular states.
Recent Publications
Ly J, Blengini CS, Cady SL, Schindler K, Cheeseman IM
Current Biology (2024)
Swartz SZ, McKay LS, Su KC, Bury L, Padeganeh A, Maddox PS, Knouse KA, Cheeseman IM
Developmental Cell (2019)
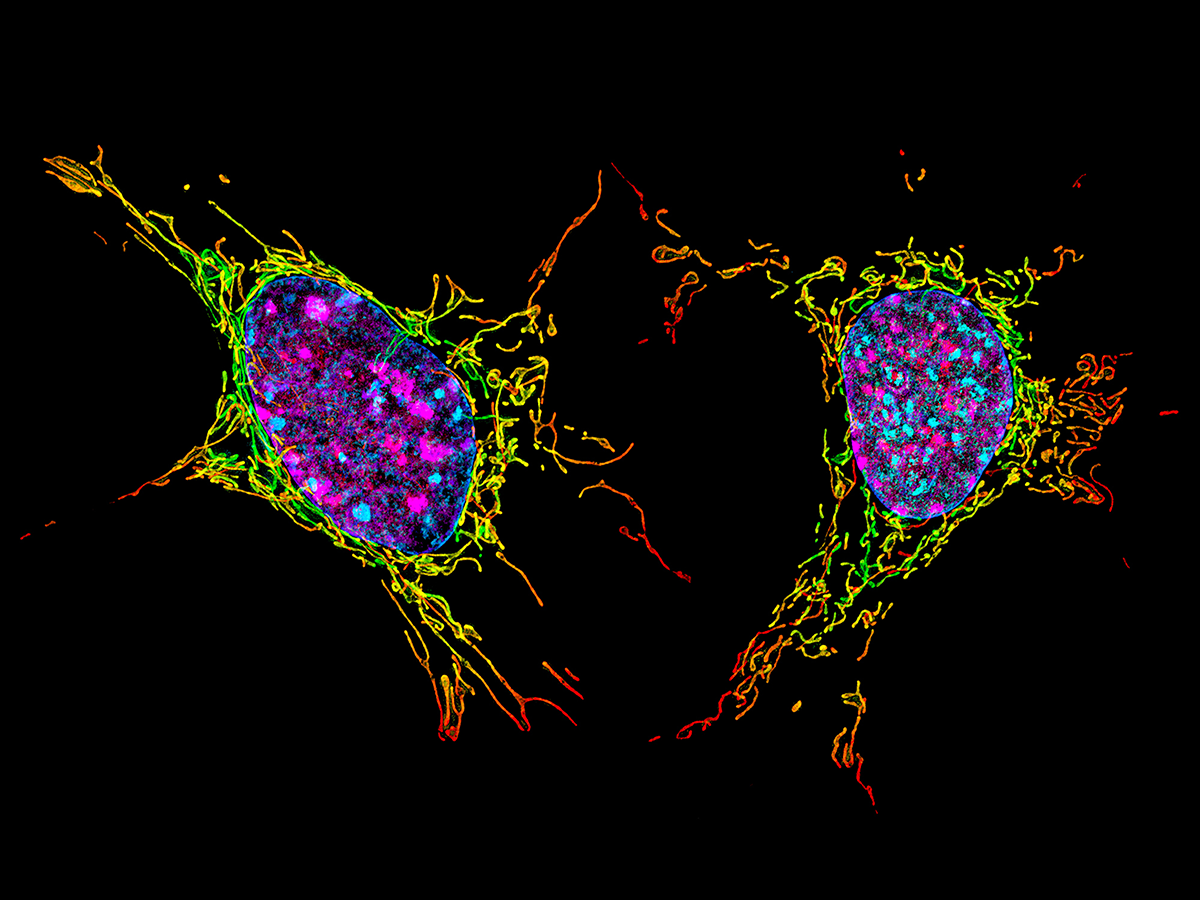
Protein Isoforms in Cell Biology and Disease
Through a combination of high-throughput ribosome profiling, functional genetics, and mass spectrometry, we have uncovered a diverse array of protein isoforms with distinct functions in healthy and disease states. Our work focuses on characterizing these previously hidden isoforms and understanding their roles in cellular processes and disease progression. We combine computational approaches with experimental validation to identify and characterize functionally- significant isoforms, particularly those relevant to cancer and rare human genetic disease. This research has revealed novel regulatory mechanisms and potential therapeutic targets across various disease contexts.
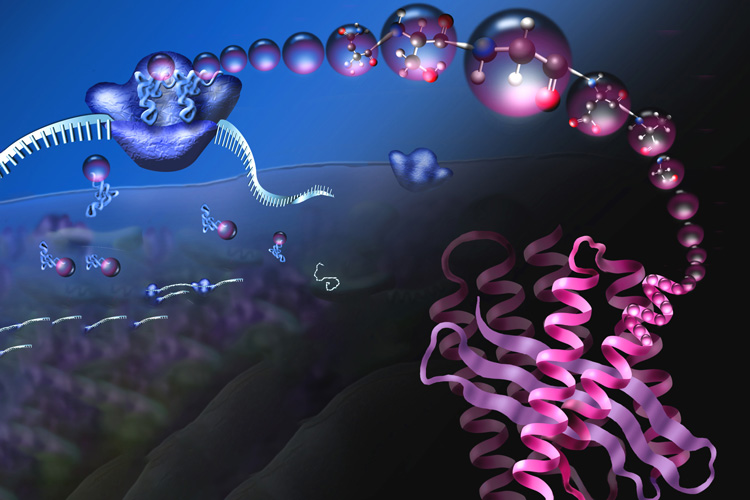
Recent Publications
Ly J, Tao YF, Di Bernardo M, Khalizeva E, Giuliano CJ, Lourido S, Fleming MD, Cheeseman IM
bioRxiv (2025)
Ly J, Xiang K, Su KC, Sissoko GB, Bartel DP, Cheeseman IM
Nature (2024)
Tsang MJ, Cheeseman IM
Nature (2023)